台大 MEG 分析處理流程
這裡是選擇sub8的exp2shortrun2_sss.fif資料進行示範
主要有四個步驟
- 根據R波位置切epoch,存成Epo.mat (其中要選比較乾淨的ECG channel)
- 去除jump和muscle artifact,存成Epo_noartifact.mat
- 做ICA,存成Epo_IC.mat
去除component,存成Epo_Rej.mat,內含4個檔案 (step 4做0~100hz的coherence,這個頻率範圍不要改)
(1) EOG與component的coherence metrix,存成eog_fdcomp.mat
(2) ECG與component的coherence metrix,存成ecg_fdcomp.mat
(3) 去除EOG與ECG的component,存成Epo_JMICA.mat
(4) 只去除EOG的component,存成Epo_JMICA_ECG.mat
第一步
流程說明
根據R波位置切epoch,存成Epo.mat (其中要選比較乾淨的ECG channel)
設定要選擇的實驗數據
subjectID=8;
condition='short'; %%%% nose , mouth , short , long
run=2; %%%% short: 1,2 long: 1,2
ID = num2str(subjectID);
R = num2str(run);載入實驗數據
這裡要注意一下
要把路徑位置改成自己本機的位置
%%% load fif file
try
%fname = sprintf(['/raid/smhsu/PA/MEG_data/sub' ID '/exp2' condition 'run' R '_sss.fif']);
if strcmp(condition,'nose') ==1
fname = sprintf(['C:/Users/king0/Documents/Matlab Code/MEG_Preprocess/sub' ID '/exp1' condition '_sss.fif']);
elseif strcmp(condition,'mouth') ==1
fname = sprintf(['C:/Users/king0/Documents/Matlab Code/MEG_Preprocess/sub' ID '/exp1' condition '_sss.fif']);
elseif strcmp(condition,'short') ==1 && run ==1
fname = sprintf(['C:/Users/king0/Documents/Matlab Code/MEG_Preprocess/sub' ID '/exp2' condition 'run' R '_sss.fif']);
elseif strcmp(condition,'short') ==1 && run ==2
fname = sprintf(['C:/Users/king0/Documents/Matlab Code/MEG_Preprocess/sub' ID '/exp2' condition 'run' R '_sss.fif']);
elseif strcmp(condition,'long') ==1 && run ==1
fname = sprintf(['C:/Users/king0/Documents/Matlab Code/MEG_Preprocess/sub' ID '/exp2' condition 'run' R '_sss.fif']);
elseif strcmp(condition,'long') ==1 && run ==2
fname = sprintf(['D:/work/pa/sub' ID '/exp2' condition 'run' R '_sss.fif']);
end
end預處理 MEG 訊號
這裡主要是將台大那邊給的訊號檔
利用ft_preprocessing進行處理的動作
%preprocessing
try
cfg = [];
cfg.dataset = fname;
data_orig = ft_preprocessing(cfg);
% ready triggers and redefine the onset and offset of the signal
event = ft_read_event(cfg.dataset);
sample_onset = [event(find(strcmp('STI001', {event.type}))).sample]';
sample_offset = [event(find(strcmp('STI002', {event.type}))).sample]';
cfg = [];
cfg.toilim = [(sample_onset+20000)/data_orig.fsample sample_offset/data_orig.fsample]; % discard the initial period, redefine a latency window in seconds
%%cfg.toilim = [1/data_orig.fsample sample_offset/data_orig.fsample];% for sub2 longrun1 only
data_new = ft_redefinetrial(cfg, data_orig);
% %%%downsample to 500hz
% cfg = [];
% cfg.resamplefs = 500;
% cfg.detrend = 'no';
% data_new_down = ft_resampledata(cfg, data_new);
end載入 R 波索引位置
%%%%%%%%load ECG R wave location
try
fname2 = ['sub' ID '_' condition '_heart_rate'];
ftpath = ['C:/Users/king0/Documents/Matlab Code/MEG_Preprocess/sub' ID ''];
load(fullfile(ftpath, fname2));
if run ==1
ECG_wave = ECG_wave1;
elseif run ==2
ECG_wave = ECG_wave2;
end
end重新轉換 ECG 訊號至原本的時間
% transform ECG_wave back to the original timeframe
ECG_wave_new = (sample_onset+20000)+ECG_wave.*2;檢查 ECG 並選擇通道
ECG 通道主要位於底下 兩個
因為 MEG 處理出來的訊號
總共有 255 個
ECG1=data_new.trial{1,1}(3,:);ECG2=data_new.trial{1,1}(4,:);
%%%check and plot, choose ECG channel
t=[180000:184000]; %%%%time (ms)
try
a=find(ECG_wave_new<=t(end) & ECG_wave_new>=t(1));
ECG1=data_new.trial{1,1}(3,:);
ECG2=data_new.trial{1,1}(4,:);
subplot(2,1,1); plot(t,ECG1(t-(sample_onset+20000))); hold on; plot(ECG_wave_new(a),ECG1(a),'rs'); xlabel('ECG003');
subplot(2,1,2); plot(t,ECG2(t-(sample_onset+20000))); hold on; plot(ECG_wave_new(a),ECG2(a),'rs'); xlabel('ECG004');
end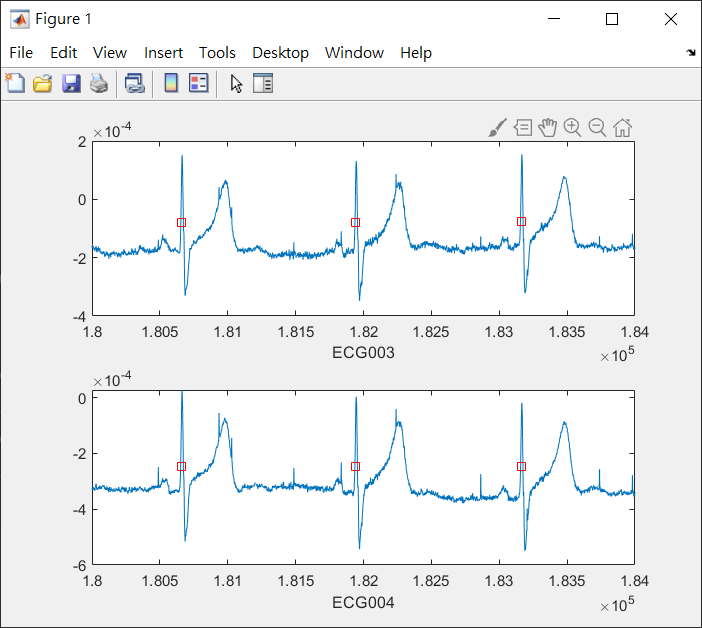
利用 ECG 的 R Peak 位置重新定義 MEG 訊號
這裡會將 MEG 訊號
透過 ECG 的 R Peak 進行切割
會分割成每一個 R Peak 就一個 MEG Data
這裡我們會定義副程式
來進行分割的動作
分割副程式
將該副程式檔名命名成trialfun_ECG.m
function [trl, event] = trialfun_ECG(cfg)
% read the header information and the events from the data
hdr = ft_read_header(cfg.dataset);
event = ft_read_event(cfg.dataset); %%%%%add
% %%%%%%%%%%%load
% if cfg.exp == 1 && cfg.condition == 1 && cfg.run ==0
% load(['sub',num2str(cfg.ID),'_nose_idRespi.mat']);
% elseif cfg.exp == 1 && cfg.condition == 2 && cfg.run ==0
% load(['sub',num2str(cfg.ID),'_mouth_idRespi.mat']);
% elseif cfg.exp == 2 && cfg.condition == 1 && cfg.run ==1
% load(['sub',num2str(cfg.ID),'_short1_idRespi.mat']);
% elseif cfg.exp == 2 && cfg.condition == 1 && cfg.run ==2
% load(['sub',num2str(cfg.ID),'_short2_idRespi.mat']);
% elseif cfg.exp == 2 && cfg.condition == 2 && cfg.run ==1
% load(['sub',num2str(cfg.ID),'_long1_idRespi.mat']);
% elseif cfg.exp == 2 && cfg.condition == 2&& cfg.run ==2
% load(['sub',num2str(cfg.ID),'_long2_idRespi.mat']);
% end
% determine the number of samples before and after the period of interest
pretrig = -round(cfg.trialdef.prestim * hdr.Fs);
posttrig = round(cfg.trialdef.poststim * hdr.Fs);
% trial epoching between two minimal points (one breath cycle)
trl = [];
for j = 1:(length(cfg.ECG_wave))
if (j+1<=length(cfg.ECG_wave)) %%%%%%%%%%%must not exceed dimension
trlbegin = cfg.ECG_wave(j+1) + pretrig; % j+1 to prevent no sufficient prestimulus period
trlend = cfg.ECG_wave(j+1) + posttrig;
offset = pretrig;
newtrl = [trlbegin trlend offset];
trl = [trl; newtrl];
end
end透過ft_definetrial來重新定義訊號
副程式寫完後
將cfg.trialfun = 'trialfun_ECG';
並透過ft_definetrial來重新定義訊號
也就是進行訊號分割的動作
%% define trial (0.4s+0.7s)
try
cfg = [];
cfg.dataset = fname;
%cfg.channel ='meg'; %%%%%%%%%
% cfg.channel ='all';
% cfg.datafile = 'data_new_down';
% cfg.headerfile = hdr;
cfg.trialfun = 'trialfun_ECG';
cfg.trialdef.prestim = 0.4;% in seconds
cfg.trialdef.poststim = 0.7; % in seconds
% cfg.ID=subjectID; %%%%%%%%%%%%%%%%%%%%%%%%%%%add ID to cfg to trialfun
% cfg.exp=exp;
% cfg.condition=condition;
% cfg.run=run;
cfg.ECG_wave=ECG_wave_new;
cfg1 = ft_definetrial(cfg);
Epo=ft_preprocessing(cfg1);
end檢查分割後訊號
這裡選擇 ECG 通道進行檢查的動作
%%%check and plot trial (check 0.4s+R wave +0.7s)
trial=145; %%%%the trial to plot
cha=2; %%%%1 or 2 (cha=1 -> ECG003 , cha=2 -> ECG004)
try
if cha==1
plot(Epo.trial{1,trial}(3,:));
elseif cha==2
plot(Epo.trial{1,trial}(4,:));
end
end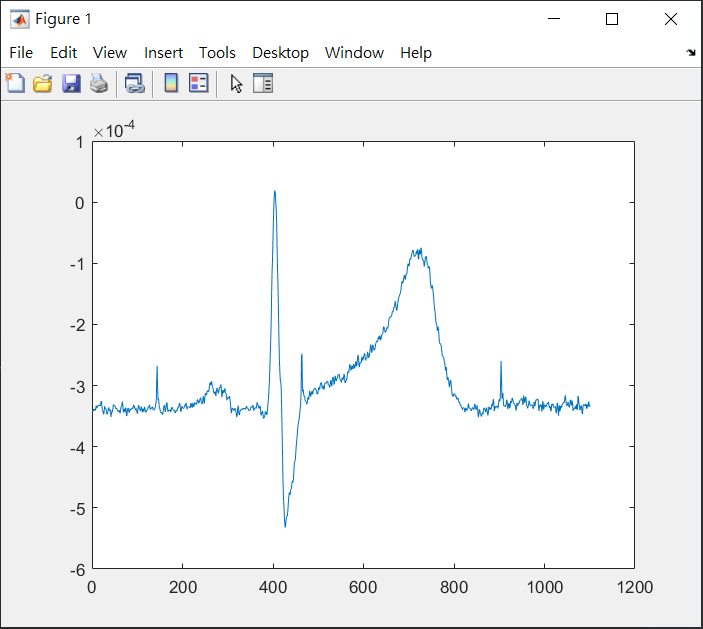
儲存檔案為 Epo 檔
%%%save
if strcmp(condition,'nose') ==1
save(['sub' ID '_' condition '_Epo'],'Epo');
elseif strcmp(condition,'mouth') ==1
save(['sub' ID '_' condition '_Epo'],'Epo');
elseif strcmp(condition,'short') ==1 && run ==1
save(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
elseif strcmp(condition,'short') ==1 && run ==2
save(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
elseif strcmp(condition,'long') ==1 && run ==1
save(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
elseif strcmp(condition,'long') ==1 && run ==2
save(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
end
%%save(['sub' ID '_' condition 'run' R '_Epo'],'Epo');第一步完整程式碼
%%ECG_step1
subjectID=8;
condition='short'; %%%% nose , mouth , short , long
run=2; %%%% short: 1,2 long: 1,2
ID = num2str(subjectID);
R = num2str(run);
%%%load fif file
try
%fname = sprintf(['/raid/smhsu/PA/MEG_data/sub' ID '/exp2' condition 'run' R '_sss.fif']);
if strcmp(condition,'nose') ==1
fname = sprintf(['C:/Users/king0/Documents/Matlab Code/MEG_Preprocess/sub' ID '/exp1' condition '_sss.fif']);
elseif strcmp(condition,'mouth') ==1
fname = sprintf(['C:/Users/king0/Documents/Matlab Code/MEG_Preprocess/sub' ID '/exp1' condition '_sss.fif']);
elseif strcmp(condition,'short') ==1 && run ==1
fname = sprintf(['C:/Users/king0/Documents/Matlab Code/MEG_Preprocess/sub' ID '/exp2' condition 'run' R '_sss.fif']);
elseif strcmp(condition,'short') ==1 && run ==2
fname = sprintf(['C:/Users/king0/Documents/Matlab Code/MEG_Preprocess/sub' ID '/exp2' condition 'run' R '_sss.fif']);
elseif strcmp(condition,'long') ==1 && run ==1
fname = sprintf(['C:/Users/king0/Documents/Matlab Code/MEG_Preprocess/sub' ID '/exp2' condition 'run' R '_sss.fif']);
elseif strcmp(condition,'long') ==1 && run ==2
fname = sprintf(['D:/work/pa/sub' ID '/exp2' condition 'run' R '_sss.fif']);
end
end
%% check ECG
%preprocessing
try
cfg = [];
cfg.dataset = fname;
data_orig = ft_preprocessing(cfg);
% ready triggers and redefine the onset and offset of the signal
event = ft_read_event(cfg.dataset);
sample_onset = [event(find(strcmp('STI001', {event.type}))).sample]';
sample_offset = [event(find(strcmp('STI002', {event.type}))).sample]';
cfg = [];
cfg.toilim = [(sample_onset+20000)/data_orig.fsample sample_offset/data_orig.fsample]; % discard the initial period, redefine a latency window in seconds
%%cfg.toilim = [1/data_orig.fsample sample_offset/data_orig.fsample];% for sub2 longrun1 only
data_new = ft_redefinetrial(cfg, data_orig);
% %%%downsample to 500hz
% cfg = [];
% cfg.resamplefs = 500;
% cfg.detrend = 'no';
% data_new_down = ft_resampledata(cfg, data_new);
end
%%%%%%%%load ECG R wave location
try
fname2 = ['sub' ID '_' condition '_heart_rate'];
ftpath = ['C:/Users/king0/Documents/Matlab Code/MEG_Preprocess/sub' ID ''];
load(fullfile(ftpath, fname2));
if run ==1
ECG_wave = ECG_wave1;
elseif run ==2
ECG_wave = ECG_wave2;
end
end
% transform ECG_wave back to the original timeframe
ECG_wave_new = (sample_onset+20000)+ECG_wave.*2;
%%%check and plot, choose ECG channel
t=[180000:184000]; %%%%time (ms)
try
a=find(ECG_wave_new<=t(end) & ECG_wave_new>=t(1));
ECG1=data_new.trial{1,1}(3,:);
ECG2=data_new.trial{1,1}(4,:);
subplot(2,1,1); plot(t,ECG1(t-(sample_onset+20000))); hold on; plot(ECG_wave_new(a),ECG1(a),'rs'); xlabel('ECG003');
subplot(2,1,2); plot(t,ECG2(t-(sample_onset+20000))); hold on; plot(ECG_wave_new(a),ECG2(a),'rs'); xlabel('ECG004');
end
%% define trial (0.4s+0.7s)
try
cfg = [];
cfg.dataset = fname;
%cfg.channel ='meg'; %%%%%%%%%
% cfg.channel ='all';
% cfg.datafile = 'data_new_down';
% cfg.headerfile = hdr;
cfg.trialfun = 'trialfun_ECG';
cfg.trialdef.prestim = 0.4;% in seconds
cfg.trialdef.poststim = 0.7; % in seconds
% cfg.ID=subjectID; %%%%%%%%%%%%%%%%%%%%%%%%%%%add ID to cfg to trialfun
% cfg.exp=exp;
% cfg.condition=condition;
% cfg.run=run;
cfg.ECG_wave=ECG_wave_new;
cfg1 = ft_definetrial(cfg);
Epo=ft_preprocessing(cfg1);
end
%%%check and plot trial (check 0.4s+R wave +0.7s)
trial=145; %%%%the trial to plot
cha=2; %%%%1 or 2 (cha=1 -> ECG003 , cha=2 -> ECG004)
try
if cha==1
plot(Epo.trial{1,trial}(3,:));
elseif cha==2
plot(Epo.trial{1,trial}(4,:));
end
end
%%%save
if strcmp(condition,'nose') ==1
save(['sub' ID '_' condition '_Epo'],'Epo');
elseif strcmp(condition,'mouth') ==1
save(['sub' ID '_' condition '_Epo'],'Epo');
elseif strcmp(condition,'short') ==1 && run ==1
save(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
elseif strcmp(condition,'short') ==1 && run ==2
save(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
elseif strcmp(condition,'long') ==1 && run ==1
save(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
elseif strcmp(condition,'long') ==1 && run ==2
save(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
end
%%save(['sub' ID '_' condition 'run' R '_Epo'],'Epo');第二步
流程說明
去除jump和muscle artifact,存成Epo_noartifact.mat
也就是進行去雜訊的動作
設定要選擇的實驗數據
%%ECG_step2
subjectID=8;
condition='short'; %%%% nose , mouth , short , long
run=2; %%%%short: 1,2 long: 1,2
ID = num2str(subjectID);
R = num2str(run);載入分割完的資料
這裡載入的是剛剛第一步分割完畢的資料
%%%load Epo
try
%load(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
if strcmp(condition,'nose') ==1
fname = sprintf(['sub' ID '_' condition '_Epo']);
load(['sub' ID '_' condition '_Epo'],'Epo');
elseif strcmp(condition,'mouth') ==1
fname = sprintf(['sub' ID '_' condition '_Epo']);
load(['sub' ID '_' condition '_Epo'],'Epo');
elseif strcmp(condition,'short') ==1 && run ==1
fname = sprintf(['sub' ID '_' condition 'run' R '_Epo']);
load(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
elseif strcmp(condition,'short') ==1 && run ==2
fname = sprintf(['sub' ID '_' condition 'run' R '_Epo']);
load(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
elseif strcmp(condition,'long') ==1 && run ==1
fname = sprintf(['sub' ID '_' condition 'run' R '_Epo']);
load(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
elseif strcmp(condition,'long') ==1 && run ==2
fname = sprintf(['sub' ID '_' condition 'run' R '_Epo']);
load(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
end
end移除 Jump 訊號
%%%%%%%%%%%% jump rejection
try
cfg = [];
%cfg.trl = trl;
% cfg.continuous = 'yes';
% channel selection, cutoff and padding
cfg.artfctdef.zvalue.channel = 'meg';
cfg.artfctdef.zvalue.cutoff = 50;
cfg.artfctdef.zvalue.trlpadding = 0;
% cfg.artfctdef.zvalue.trlpadding = -0.2;
cfg.artfctdef.zvalue.artpadding = 0;
cfg.artfctdef.zvalue.fltpadding = 0;
% algorithmic parameters
cfg.artfctdef.zvalue.cumulative = 'yes';
cfg.artfctdef.zvalue.medianfilter = 'yes';
cfg.artfctdef.zvalue.medianfiltord = 9;
cfg.artfctdef.zvalue.absdiff = 'yes';
% make the process interactive
cfg.artfctdef.zvalue.interactive = 'yes';
[cfg, Epo_J] = ft_artifact_zvalue(cfg, Epo);
end該程式碼執行過後
會出現以圖形介面的方式
讓你進行移除訊號的動作
選擇欲移除的訊號片段
點擊reject full後 關閉該頁面
即可移除該訊號
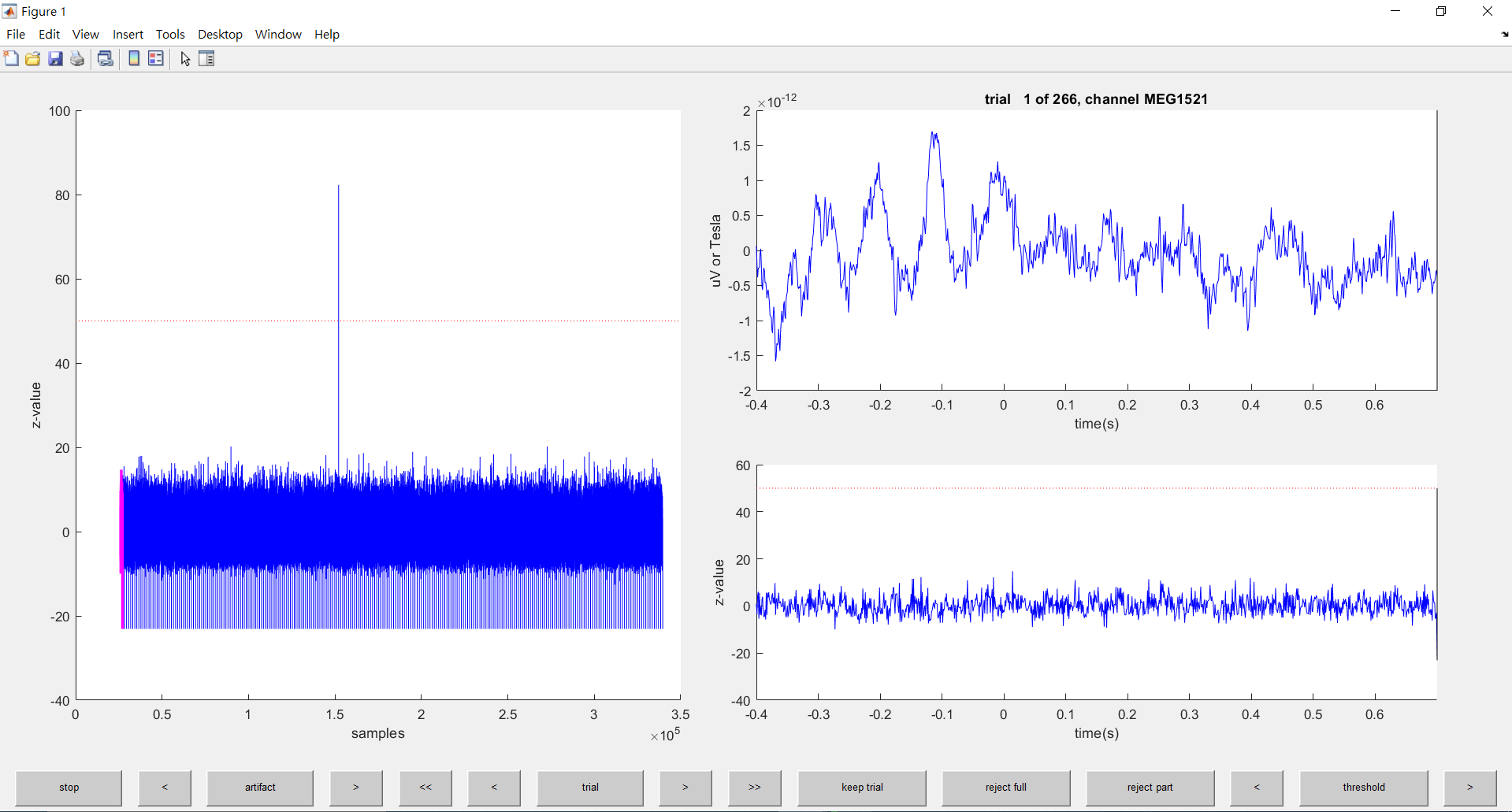
移除肌肉跳動的訊號
%%%%% muscle rejection
try
% muscle
cfg = [];
%cfg.trl = trl;
% cfg.continuous = 'yes';
% channel selection, cutoff and padding
cfg.artfctdef.zvalue.channel = 'meg';
cfg.artfctdef.zvalue.cutoff = 50;
cfg.artfctdef.zvalue.trlpadding = 0;
% cfg.artfctdef.zvalue.trlpadding = -0.2;
cfg.artfctdef.zvalue.fltpadding = 0;
% cfg.artfctdef.zvalue.artpadding = 0.1;
cfg.artfctdef.zvalue.artpadding = 0;
% algorithmic parameters
cfg.artfctdef.zvalue.bpfilter = 'yes';
cfg.artfctdef.zvalue.bpfreq = [110 140];
cfg.artfctdef.zvalue.bpfiltord = 9;
cfg.artfctdef.zvalue.bpfilttype = 'but';
cfg.artfctdef.zvalue.hilbert = 'yes';
cfg.artfctdef.zvalue.boxcar = 0.2;
% make the process interactive
cfg.artfctdef.zvalue.interactive = 'yes';
[cfg, Epo_M] = ft_artifact_zvalue(cfg, Epo);
end這一步跟上一步一樣
選擇你覺得像雜訊的波型
進行移除就行了
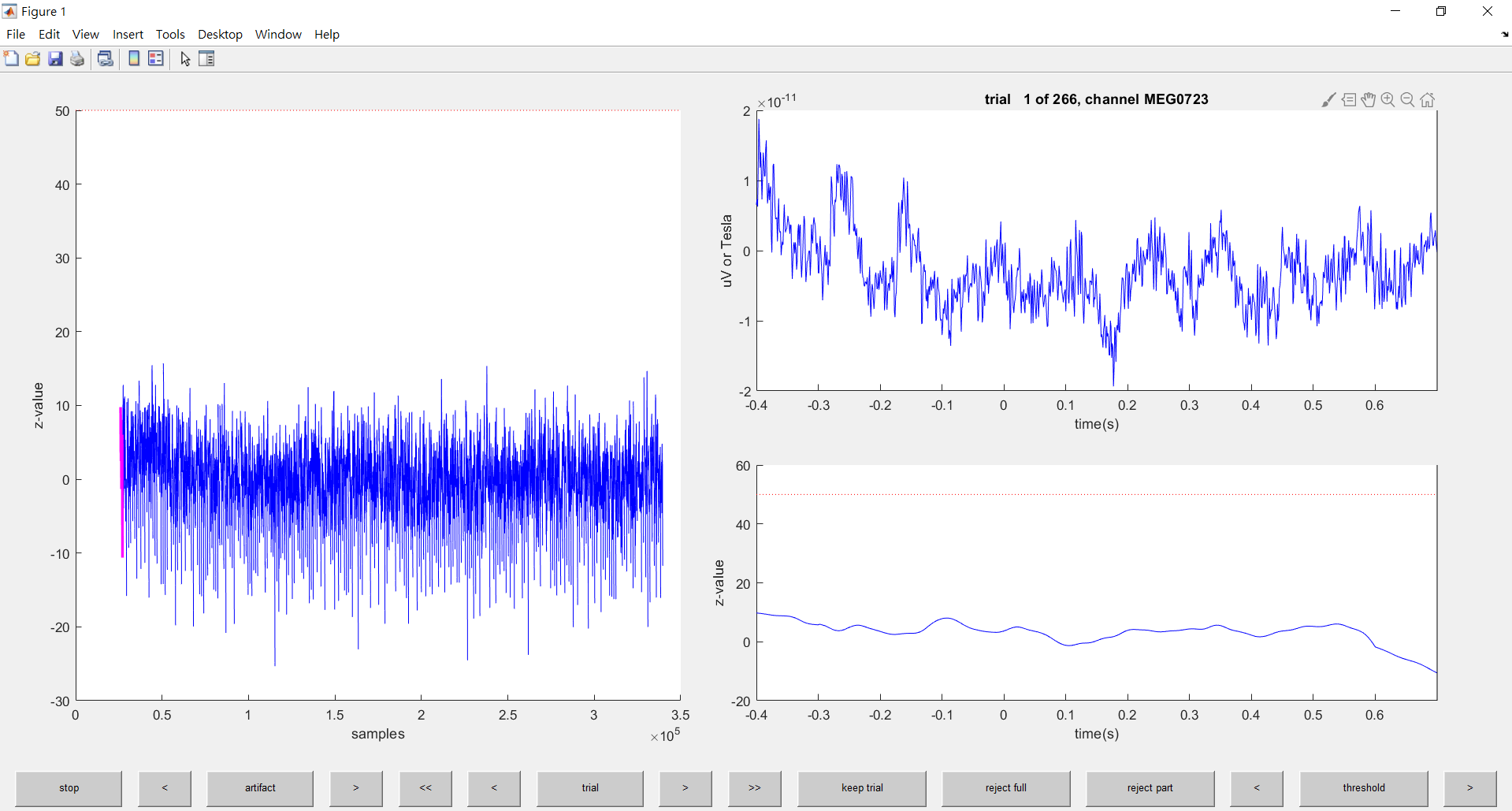
移除 jump 和 muscle
因為前兩步 是將雜訊位置儲存成變數
所以才是正式將原訊號進行移除雜訊的動作
透過artfctdef進行去除的動作
更詳細的方法可以參考官網
artfctdef: http://www.fieldtriptoolbox.org/reference/ft_rejectartifact/
%%%%%reject jump and muscle
try
cfg=[];
% cfg.artfctdef.reject = 'partial';
cfg.artfctdef.jump.artifact = Epo_J;
cfg.artfctdef.muscle.artifact = Epo_M;
Epo_no_artifacts = ft_rejectartifact(cfg,Epo);
end
%當用圖形介面去除 jump 和 muscle 失敗時的方法
%%%%%%when jump and muscle can't work, %%%%%%%%%%%%(very important)
Epo_no_artifacts=Epo;
%%%%%only keep ECG channel
try
cfg = [];
cfg.channel = {'ECG003','ECG004'}; %MISC(respiration),STI001(exp onset), STI002(exp offset)
Epo_temp = ft_preprocessing(cfg,Epo_no_artifacts);
end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%檢查訊號全部分割訊號
%%%%%%%%%%%%%%%%%%%check trial
cha=2; %%%the same as step 1
%%%press > or < button to ECG003 or ECG004 channel to check every trial
try
% cfg=[];
% cfg.method = 'trial';
% if cha==1
% cfg.channel = {'ECG003'};
% end
% if cha==2
% cfg.channel = {'ECG004'};
% end
cfg=[];
cfg.method = 'channel';
ft_rejectvisual(cfg, Epo_no_artifacts)
end
%這裡可以讓你概觀的檢查所有的分割訊號
如果你有不滿意的訊號
你可以紀錄 該訊號的編號
後面會寫個程式 來將其進行刪除的動作
刪除訊號片段
在delete_trial裡面的陣列
填寫欲刪除的編號
%%%delete trial
delete_trial=[127];
try
trialsToKeep = 1:length(Epo_no_artifacts.trial);
trialsToKeep(delete_trial) = [];
cfg = [];
cfg.trials = trialsToKeep;
Epo_no_artifacts = ft_preprocessing(cfg, Epo_no_artifacts);
end
%儲存檔案
%%%save
if strcmp(condition,'nose') ==1
save(['sub' ID '_' condition '_Epo_noartifact'],'Epo_no_artifacts');
elseif strcmp(condition,'mouth') ==1
save(['sub' ID '_' condition '_Epo_noartifact'],'Epo_no_artifacts');
elseif strcmp(condition,'short') ==1 && run ==1
save(['sub' ID '_' condition 'run' R '_Epo_noartifact'],'Epo_no_artifacts');
elseif strcmp(condition,'short') ==1 && run ==2
save(['sub' ID '_' condition 'run' R '_Epo_noartifact'],'Epo_no_artifacts');
elseif strcmp(condition,'long') ==1 && run ==1
save(['sub' ID '_' condition 'run' R '_Epo_noartifact'],'Epo_no_artifacts');
elseif strcmp(condition,'long') ==1 && run ==2
save(['sub' ID '_' condition 'run' R '_Epo_noartifact'],'Epo_no_artifacts');
end完整程式碼
%%ECG_step2
subjectID=8;
condition='short'; %%%% nose , mouth , short , long
run=2; %%%%short: 1,2 long: 1,2
ID = num2str(subjectID);
R = num2str(run);
%%%load Epo
try
%load(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
if strcmp(condition,'nose') ==1
fname = sprintf(['sub' ID '_' condition '_Epo']);
load(['sub' ID '_' condition '_Epo'],'Epo');
elseif strcmp(condition,'mouth') ==1
fname = sprintf(['sub' ID '_' condition '_Epo']);
load(['sub' ID '_' condition '_Epo'],'Epo');
elseif strcmp(condition,'short') ==1 && run ==1
fname = sprintf(['sub' ID '_' condition 'run' R '_Epo']);
load(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
elseif strcmp(condition,'short') ==1 && run ==2
fname = sprintf(['sub' ID '_' condition 'run' R '_Epo']);
load(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
elseif strcmp(condition,'long') ==1 && run ==1
fname = sprintf(['sub' ID '_' condition 'run' R '_Epo']);
load(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
elseif strcmp(condition,'long') ==1 && run ==2
fname = sprintf(['sub' ID '_' condition 'run' R '_Epo']);
load(['sub' ID '_' condition 'run' R '_Epo'],'Epo');
end
end
%%%%%%%%%%%% jump rejection
try
cfg = [];
%cfg.trl = trl;
% cfg.continuous = 'yes';
% channel selection, cutoff and padding
cfg.artfctdef.zvalue.channel = 'meg';
cfg.artfctdef.zvalue.cutoff = 50;
cfg.artfctdef.zvalue.trlpadding = 0;
% cfg.artfctdef.zvalue.trlpadding = -0.2;
cfg.artfctdef.zvalue.artpadding = 0;
cfg.artfctdef.zvalue.fltpadding = 0;
% algorithmic parameters
cfg.artfctdef.zvalue.cumulative = 'yes';
cfg.artfctdef.zvalue.medianfilter = 'yes';
cfg.artfctdef.zvalue.medianfiltord = 9;
cfg.artfctdef.zvalue.absdiff = 'yes';
% make the process interactive
cfg.artfctdef.zvalue.interactive = 'yes';
[cfg, Epo_J] = ft_artifact_zvalue(cfg, Epo);
end
%%%%% muscle rejection
try
% muscle
cfg = [];
%cfg.trl = trl;
% cfg.continuous = 'yes';
% channel selection, cutoff and padding
cfg.artfctdef.zvalue.channel = 'meg';
cfg.artfctdef.zvalue.cutoff = 50;
cfg.artfctdef.zvalue.trlpadding = 0;
% cfg.artfctdef.zvalue.trlpadding = -0.2;
cfg.artfctdef.zvalue.fltpadding = 0;
% cfg.artfctdef.zvalue.artpadding = 0.1;
cfg.artfctdef.zvalue.artpadding = 0;
% algorithmic parameters
cfg.artfctdef.zvalue.bpfilter = 'yes';
cfg.artfctdef.zvalue.bpfreq = [110 140];
cfg.artfctdef.zvalue.bpfiltord = 9;
cfg.artfctdef.zvalue.bpfilttype = 'but';
cfg.artfctdef.zvalue.hilbert = 'yes';
cfg.artfctdef.zvalue.boxcar = 0.2;
% make the process interactive
cfg.artfctdef.zvalue.interactive = 'yes';
[cfg, Epo_M] = ft_artifact_zvalue(cfg, Epo);
end
%
%%%%%reject jump and muscle
try
cfg=[];
% cfg.artfctdef.reject = 'partial';
cfg.artfctdef.jump.artifact = Epo_J;
cfg.artfctdef.muscle.artifact = Epo_M;
Epo_no_artifacts = ft_rejectartifact(cfg,Epo);
end
%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%when jump and muscle can't work, %%%%%%%%%%%%(very important)
Epo_no_artifacts=Epo;
%%%%%only keep ECG channel
try
cfg = [];
cfg.channel = {'ECG003','ECG004'}; %MISC(respiration),STI001(exp onset), STI002(exp offset)
Epo_temp = ft_preprocessing(cfg,Epo_no_artifacts);
end
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%%%%%%%%%%%%%%%%%%%check trial
cha=2; %%%the same as step 1
%%%press > or < button to ECG003 or ECG004 channel to check every trial
try
% cfg=[];
% cfg.method = 'trial';
% if cha==1
% cfg.channel = {'ECG003'};
% end
% if cha==2
% cfg.channel = {'ECG004'};
% end
cfg=[];
cfg.method = 'channel';
ft_rejectvisual(cfg, Epo_no_artifacts)
end
%
%%%delete trial
delete_trial=[127];
try
trialsToKeep = 1:length(Epo_no_artifacts.trial);
trialsToKeep(delete_trial) = [];
cfg = [];
cfg.trials = trialsToKeep;
Epo_no_artifacts = ft_preprocessing(cfg, Epo_no_artifacts);
end
%
%%%save
if strcmp(condition,'nose') ==1
save(['sub' ID '_' condition '_Epo_noartifact'],'Epo_no_artifacts');
elseif strcmp(condition,'mouth') ==1
save(['sub' ID '_' condition '_Epo_noartifact'],'Epo_no_artifacts');
elseif strcmp(condition,'short') ==1 && run ==1
save(['sub' ID '_' condition 'run' R '_Epo_noartifact'],'Epo_no_artifacts');
elseif strcmp(condition,'short') ==1 && run ==2
save(['sub' ID '_' condition 'run' R '_Epo_noartifact'],'Epo_no_artifacts');
elseif strcmp(condition,'long') ==1 && run ==1
save(['sub' ID '_' condition 'run' R '_Epo_noartifact'],'Epo_no_artifacts');
elseif strcmp(condition,'long') ==1 && run ==2
save(['sub' ID '_' condition 'run' R '_Epo_noartifact'],'Epo_no_artifacts');
end
%%%save(['sub' ID '_' condition 'run' R '_Epo_noartifact'],'Epo_no_artifacts');第三步
選擇資料
%%ECG_step2
subjectID=8;
condition='short'; %%%% nose , mouth , short , long
run=2; %%%%short: 1,2 long: 1,2
ID = num2str(subjectID);
R = num2str(run);載入資料
%%%load Epo_noartifact
try
%load(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
if strcmp(condition,'nose') ==1
fname = sprintf(['sub' ID '_' condition '_Epo_noartifact']);
load(['sub' ID '_' condition '_Epo_noartifact']);
elseif strcmp(condition,'mouth') ==1
fname = sprintf(['sub' ID '_' condition '_Epo_noartifact']);
load(['sub' ID '_' condition '_Epo_noartifact']);
elseif strcmp(condition,'short') ==1 && run ==1
fname = sprintf(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
load(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
elseif strcmp(condition,'short') ==1 && run ==2
fname = sprintf(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
load(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
elseif strcmp(condition,'long') ==1 && run ==1
fname = sprintf(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
load(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
elseif strcmp(condition,'long') ==1 && run ==2
fname = sprintf(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
load(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
end
endICA 獨立成分分析
這裡主要將訊號重新取樣至 500
而我們在進行ICA前 需先設定 n_comp 參數
n_comp 參數為該訊號的個數或維度
%%%%%%%%%%%%% ICA
%%% take meg to compute n_comp,downsample
try
%%%% take meg to compute n_comp
cfg=[];
cfg.channel = 'meg';
Epo_meg = ft_preprocessing(cfg,Epo_no_artifacts);
n_comp = rank(squeeze(Epo_meg.trial{1}) * squeeze(Epo_meg.trial{1})');
%%%%%%% downsample
cfg = [];
cfg.resamplefs = 500;
cfg.detrend = 'no';
Epo_no_artifacts_down = ft_resampledata(cfg, Epo_no_artifacts);
end
%獨立成分分析
% perform the independent component analysis (i.e., decompose the data)
try
cfg = [];
cfg.channel = 'meg';
cfg.method = 'runica'; % this is the default and uses the implementation from EEGLAB
%n_comp = rank(squeeze(Epo_no_artifacts_down.trial{1}) * squeeze(Epo_no_artifacts_down.trial{1})');
n_comp = rank(squeeze(Epo_meg.trial{1}) * squeeze(Epo_meg.trial{1})');
cfg.runica.pca = n_comp;
cfg.runica.stop = 1e-7;
comp = ft_componentanalysis(cfg, Epo_no_artifacts_down);
end
%儲存檔案
%%%save comp
if strcmp(condition,'nose') ==1
save(['sub' ID '_' condition '_Epo_IC'],'comp');
elseif strcmp(condition,'mouth') ==1
save(['sub' ID '_' condition '_Epo_IC'],'comp');
elseif strcmp(condition,'short') ==1 && run ==1
save(['sub' ID '_' condition 'run' R '_Epo_IC'],'comp');
elseif strcmp(condition,'short') ==1 && run ==2
save(['sub' ID '_' condition 'run' R '_Epo_IC'],'comp');
elseif strcmp(condition,'long') ==1 && run ==1
save(['sub' ID '_' condition 'run' R '_Epo_IC'],'comp');
elseif strcmp(condition,'long') ==1 && run ==2
save(['sub' ID '_' condition 'run' R '_Epo_IC'],'comp');
end
%%save(['sub' ID '_' condition 'run' R '_Epo_IC'],'comp');完整程式碼
%%ECG_step2
subjectID=8;
condition='short'; %%%% nose , mouth , short , long
run=2; %%%%short: 1,2 long: 1,2
ID = num2str(subjectID);
R = num2str(run);
%%%load Epo_noartifact
try
%load(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
if strcmp(condition,'nose') ==1
fname = sprintf(['sub' ID '_' condition '_Epo_noartifact']);
load(['sub' ID '_' condition '_Epo_noartifact']);
elseif strcmp(condition,'mouth') ==1
fname = sprintf(['sub' ID '_' condition '_Epo_noartifact']);
load(['sub' ID '_' condition '_Epo_noartifact']);
elseif strcmp(condition,'short') ==1 && run ==1
fname = sprintf(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
load(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
elseif strcmp(condition,'short') ==1 && run ==2
fname = sprintf(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
load(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
elseif strcmp(condition,'long') ==1 && run ==1
fname = sprintf(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
load(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
elseif strcmp(condition,'long') ==1 && run ==2
fname = sprintf(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
load(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
end
end
%%%%%%%%%%%%% ICA
%%%take meg to compute n_comp,downsample
try
%%%%take meg to compute n_comp
cfg=[];
cfg.channel = 'meg';
Epo_meg = ft_preprocessing(cfg,Epo_no_artifacts);
n_comp = rank(squeeze(Epo_meg.trial{1}) * squeeze(Epo_meg.trial{1})');
%%%%%%%downsample
cfg = [];
cfg.resamplefs = 500;
cfg.detrend = 'no';
Epo_no_artifacts_down = ft_resampledata(cfg, Epo_no_artifacts);
end
%
% perform the independent component analysis (i.e., decompose the data)
try
cfg = [];
cfg.channel = 'meg';
cfg.method = 'runica'; % this is the default and uses the implementation from EEGLAB
%n_comp = rank(squeeze(Epo_no_artifacts_down.trial{1}) * squeeze(Epo_no_artifacts_down.trial{1})');
n_comp = rank(squeeze(Epo_meg.trial{1}) * squeeze(Epo_meg.trial{1})');
cfg.runica.pca = n_comp;
cfg.runica.stop = 1e-7;
comp = ft_componentanalysis(cfg, Epo_no_artifacts_down);
end
%
%%%save comp
if strcmp(condition,'nose') ==1
save(['sub' ID '_' condition '_Epo_IC'],'comp');
elseif strcmp(condition,'mouth') ==1
save(['sub' ID '_' condition '_Epo_IC'],'comp');
elseif strcmp(condition,'short') ==1 && run ==1
save(['sub' ID '_' condition 'run' R '_Epo_IC'],'comp');
elseif strcmp(condition,'short') ==1 && run ==2
save(['sub' ID '_' condition 'run' R '_Epo_IC'],'comp');
elseif strcmp(condition,'long') ==1 && run ==1
save(['sub' ID '_' condition 'run' R '_Epo_IC'],'comp');
elseif strcmp(condition,'long') ==1 && run ==2
save(['sub' ID '_' condition 'run' R '_Epo_IC'],'comp');
end
%%save(['sub' ID '_' condition 'run' R '_Epo_IC'],'comp');第四步
選擇資料
subjectID=8;
condition='short'; %%%% nose , mouth , short , long
run=2; %%%%short: 1,2 long: 1,2
ID = num2str(subjectID);
R = num2str(run);載入資料
這裡主要是載入
去除雜訊、做完ICA、原始訊號檔
%%%load Epo_noarttifact and Epo_IC and fif file
try
%load(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
%load(['sub' ID '_' condition 'run' R '_Epo_IC']);
if strcmp(condition,'nose') ==1
fname = sprintf(['D:/work/pa/sub' ID '/exp1' condition '_sss.fif']);
load(['sub' ID '_' condition '_Epo_noartifact']);
load(['sub' ID '_' condition '_Epo_IC']);
elseif strcmp(condition,'mouth') ==1
fname = sprintf(['D:/work/pa/sub' ID '/exp1' condition '_sss.fif']);
load(['sub' ID '_' condition '_Epo_noartifact']);
load(['sub' ID '_' condition '_Epo_IC']);
elseif strcmp(condition,'short') ==1 && run ==1
fname = sprintf(['D:/work/pa/sub' ID '/exp2' condition 'run' R '_sss.fif']);
load(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
load(['sub' ID '_' condition 'run' R '_Epo_IC']);
elseif strcmp(condition,'short') ==1 && run ==2
fname = sprintf(['D:/work/pa/sub' ID '/exp2' condition 'run' R '_sss.fif']);
load(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
load(['sub' ID '_' condition 'run' R '_Epo_IC']);
elseif strcmp(condition,'long') ==1 && run ==1
fname = sprintf(['D:/work/pa/sub' ID '/exp2' condition 'run' R '_sss.fif']);
load(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
load(['sub' ID '_' condition 'run' R '_Epo_IC']);
elseif strcmp(condition,'long') ==1 && run ==2
fname = sprintf(['D:/work/pa/sub' ID '/exp2' condition 'run' R '_sss.fif']);
load(['sub' ID '_' condition 'run' R '_Epo_noartifact']);
load(['sub' ID '_' condition 'run' R '_Epo_IC']);
end
end畫出腦磁圖
%%%%%%plot the components for visual inspection (modify or default) (picture)
figure;
cfg = [];
cfg.component = [1:30]; %(modify or default) specify the component(s) that should be plotted
cfg.layout = 'neuromag306all.lay'; % specify the layout file that should be used for plotting
cfg.comment = 'no';
ft_topoplotIC(cfg, comp)
重新處理訊號
%%%data
try
cfg = [];
cfg.dataset = fname;
cfg.channel = {'MISC001','STI*','ecg','eog','emg'};%MISC(respiration),STI001(exp onset), STI002(exp offset)
data_orig = ft_preprocessing(cfg);
% ready triggers and redefine the onset and offset of the signal
event = ft_read_event(cfg.dataset);
sample_onset = [event(find(strcmp('STI001', {event.type}))).sample]';
sample_offset = [event(find(strcmp('STI002', {event.type}))).sample]';
cfg = [];
cfg.toilim = [(sample_onset+20000)/data_orig.fsample sample_offset/data_orig.fsample]; % discard the initial period, redefine a latency window in seconds
data_new = ft_redefinetrial(cfg, data_orig);
end
%畫出磁強計
%%%%%%visualization of magnetometers (auto) (picture)
try
cfg = [];
cfg.layout = 'neuromag306mag.lay'; % specify the layout file that should be used for plotting
cfg.viewmode = 'component';
ft_databrowser(cfg, comp)
end
%
檢查訊號
這裡可以自行選擇片段來進行檢查
可以檢查 EOG EOG ECG ECG ECG
%%%choose trial to check
trial=14; %%%(modify)
try
figure;
a=Epo_no_artifacts.sampleinfo(trial,1)-data_new.sampleinfo(1)+1;
b=Epo_no_artifacts.sampleinfo(trial,2)-data_new.sampleinfo(1)+1;
subplot(4,1,1), plot(data_new.trial{1,1}(1,a:b)); title('EOG'); %%%%%EOG EOG ECG ECG ECG
subplot(4,1,2), plot(data_new.trial{1,1}(2,a:b)); title('EOG');
subplot(4,1,3), plot(data_new.trial{1,1}(3,a:b)); title('ECG');
subplot(4,1,4), plot(data_new.trial{1,1}(4,a:b)); title('ECG');
end
%
畫出 gradiometers
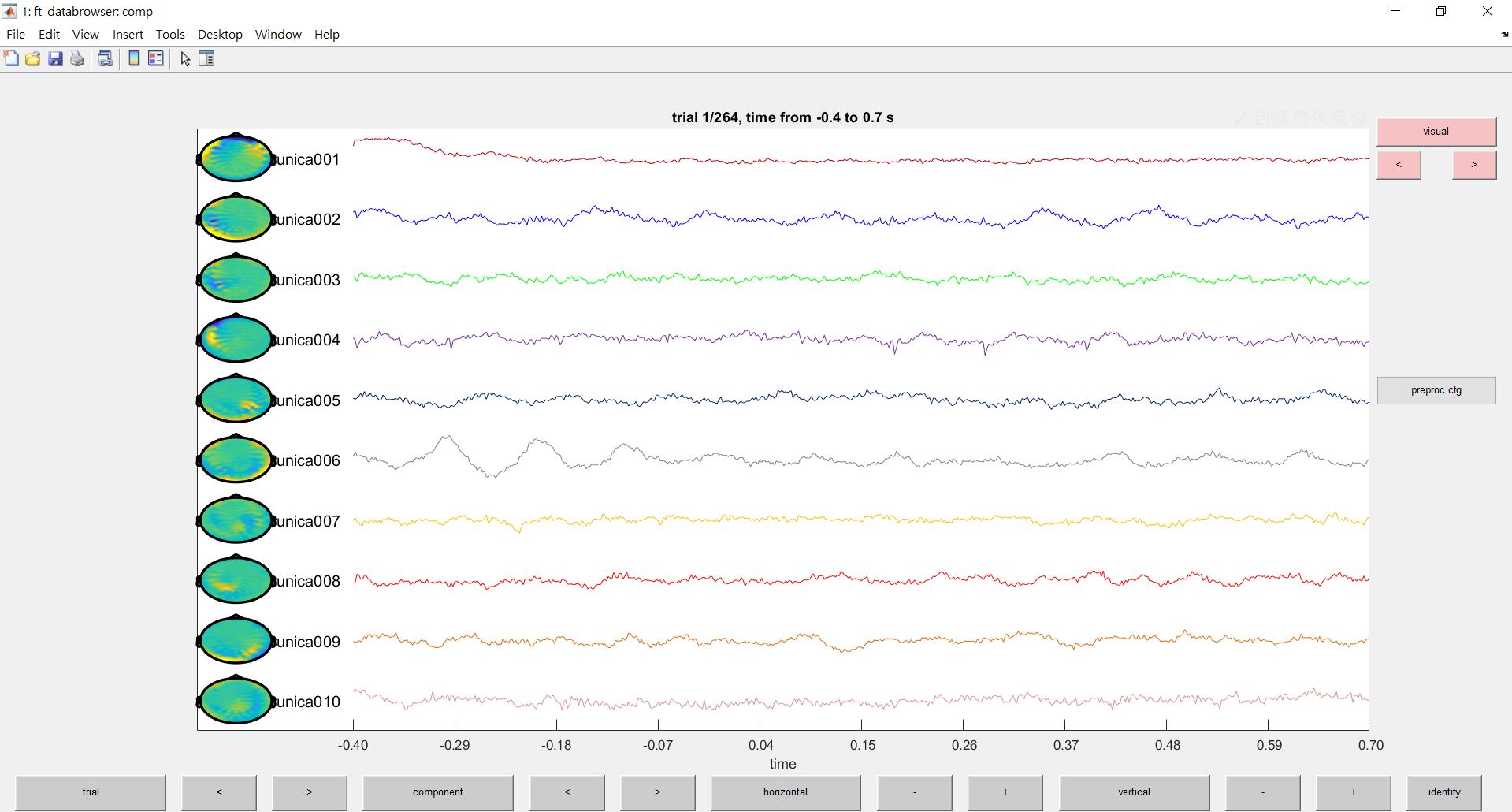
%%%%%%visualization of gradiometers (auto) (picture)
try
cfg = [];
cfg.layout = 'neuromag306planar.lay'; % specify the layout file that should be used for plotting
cfg.viewmode = 'component';
ft_databrowser(cfg, comp)
end
%
compute coherence between all components and the EOG (around 0~100hz)
%%%%compute coherence between all components and the EOG (around 0~100hz)
cha=2; %%%%1 or 2 (cha=1 -> EOG001 , cha=2 -> EOG002)
try
cfg=[];
if cha==1
cfg.channel = {'EOG001'};
elseif cha==2
cfg.channel = {'EOG002'};
end
eog = ft_preprocessing(cfg,Epo_no_artifacts);
%%%%downsample
cfg = [];
cfg.resamplefs = 500;
cfg.detrend = 'no';
eog_down = ft_resampledata(cfg, eog);
% append the eog channel to the data structure
comp_eog = ft_appenddata([], eog_down, comp);
% compute a frequency decomposition of all components and the EOG
cfg = [];
cfg.method = 'mtmfft';
cfg.output = 'fourier';
cfg.foilim = [0 100]; %%%%%%%%%hz
cfg.taper = 'hanning';
cfg.pad = 'maxperlen';
freq = ft_freqanalysis(cfg, comp_eog);
% compute coherence between all components and the EOG
cfg = [];
cfg.channelcmb = {'all' 'eog'};
cfg.jackknife = 'no';
cfg.method = 'coh';
eog_fdcomp = ft_connectivityanalysis(cfg, freq);
end
%look at the coherence spectrum between all components and the EOG
figure;
subplot(2,1,1); plot(eog_fdcomp.freq, abs(eog_fdcomp.cohspctrm)); xlabel('Hz'); ylabel('coherence');
subplot(2,1,2); imagesc(eog_fdcomp.freq,[1:length(eog_fdcomp.labelcmb)],abs(eog_fdcomp.cohspctrm)); xlabel('Hz'); ylabel('component');
eog_comp_avg=mean(eog_fdcomp.cohspctrm(:,7),2); %%%0~5.44hz avg
[value,probabaly_eog_comp]=max(eog_comp_avg) %%%find component of max value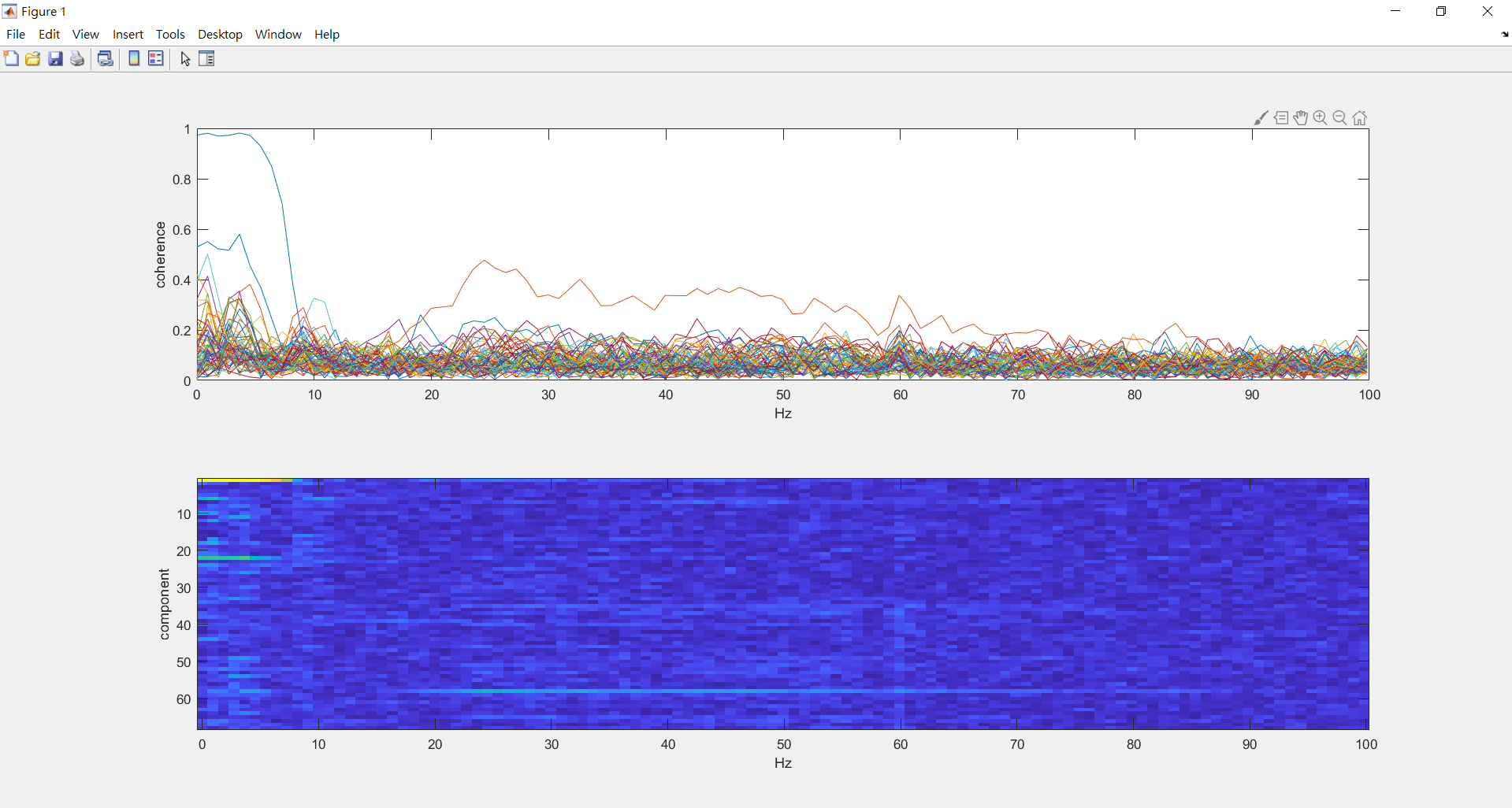
compute coherence between all components and the ECG (around 0~100hz)
%%%%compute coherence between all components and the ECG (around 0~100hz)
cha=2; %%%the same as step 1
try
cfg=[];
if cha==1
cfg.channel = {'ECG003'};
elseif cha==2
cfg.channel = {'ECG004'};
end
ecg = ft_preprocessing(cfg,Epo_no_artifacts);
%%%%downsample
cfg = [];
cfg.resamplefs = 500;
cfg.detrend = 'no';
ecg_down = ft_resampledata(cfg, ecg);
% append the ecg channel to the data structure
comp_ecg = ft_appenddata([], ecg_down, comp);
% compute a frequency decomposition of all components and the ECG
cfg = [];
cfg.method = 'mtmfft';
cfg.output = 'fourier';
cfg.foilim = [0 100]; %%%%hz
cfg.taper = 'hanning';
cfg.pad = 'maxperlen';
freq = ft_freqanalysis(cfg, comp_ecg);
% compute coherence between all components and the ECG
cfg = [];
cfg.channelcmb = {'all' 'ecg'};
cfg.jackknife = 'no';
cfg.method = 'coh';
ecg_fdcomp = ft_connectivityanalysis(cfg, freq);
end
%look at the coherence spectrum between all components and the ECG
figure;
subplot(2,1,1); plot(ecg_fdcomp.freq, abs(ecg_fdcomp.cohspctrm)); xlabel('Hz'); ylabel('coherence');
subplot(2,1,2); imagesc(ecg_fdcomp.freq,[1:length(ecg_fdcomp.labelcmb)],abs(ecg_fdcomp.cohspctrm)); xlabel('Hz'); ylabel('component');
ecg_comp_avg=mean(ecg_fdcomp.cohspctrm(:,35),2); %%%0~30.85hz avg
[value,probabaly_ecg_comp]=max(ecg_comp_avg) %%%find component of max value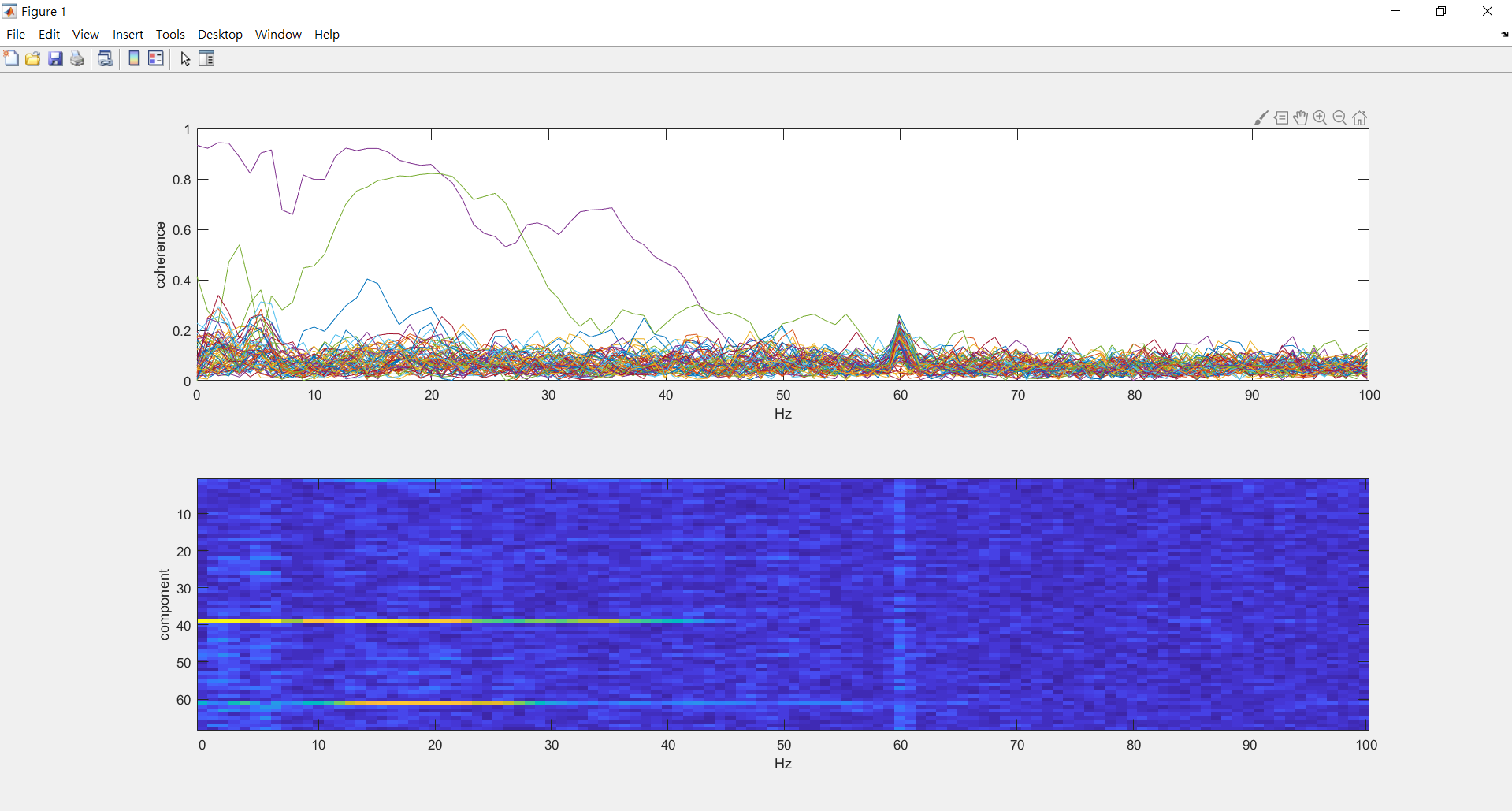
刪除 EOG 和 ECG component
%%%%%%delete EOG and ECG component (modify) (very important)%%%%%%%%%
comp_delete=[1 37]; %(modify)modify which component to delete (small to big)
try
%%%%%%%downsample
cfg = [];
cfg.resamplefs = 500;
cfg.detrend = 'no';
Epo_no_artifacts_down = ft_resampledata(cfg, Epo_no_artifacts);
%%%delete
cfg = [];
cfg.component = comp_delete; %%
Epo_JMICA = ft_rejectcomponent(cfg, comp, Epo_no_artifacts_down);
end
%
%%%%%%only delete EOG component (modify) (very important)%%%%%%%%%
comp_delete=[1]; %(modify)modify which component to delete (small to big)
try
%%%%%%%downsample
cfg = [];
cfg.resamplefs = 500;
cfg.detrend = 'no';
Epo_no_artifacts_down = ft_resampledata(cfg, Epo_no_artifacts);
%%%delete
cfg = [];
cfg.component = comp_delete; %%
Epo_JMICA_ECG = ft_rejectcomponent(cfg, comp, Epo_no_artifacts_down);
end
%儲存Epo、EOG coherence、 ECG coherence
%%%%save Epo, EOG coherence, ECG coherence
if strcmp(condition,'nose') ==1
save(['sub' ID '_' condition '_Epo_Rej'],'Epo_JMICA','Epo_JMICA_ECG','eog_fdcomp','ecg_fdcomp');
elseif strcmp(condition,'mouth') ==1
save(['sub' ID '_' condition '_Epo_Rej'],'Epo_JMICA','Epo_JMICA_ECG','eog_fdcomp','ecg_fdcomp');
elseif strcmp(condition,'short') ==1 && run ==1
save(['sub' ID '_' condition 'run' R '_Epo_Rej'],'Epo_JMICA','Epo_JMICA_ECG','eog_fdcomp','ecg_fdcomp');
elseif strcmp(condition,'short') ==1 && run ==2
save(['sub' ID '_' condition 'run' R '_Epo_Rej'],'Epo_JMICA','Epo_JMICA_ECG','eog_fdcomp','ecg_fdcomp');
elseif strcmp(condition,'long') ==1 && run ==1
save(['sub' ID '_' condition 'run' R '_Epo_Rej'],'Epo_JMICA','Epo_JMICA_ECG','eog_fdcomp','ecg_fdcomp');
elseif strcmp(condition,'long') ==1 && run ==2
save(['sub' ID '_' condition 'run' R '_Epo_Rej'],'Epo_JMICA','Epo_JMICA_ECG','eog_fdcomp','ecg_fdcomp');
end
One comment
毅鈞:
你之前的碩士論文結果將先投到FENS forum 2026, 能否將你現今的email留給我(徐博士)一下,謝謝!